

|
|
|||||
|
|
|||||
|
|
Analysis of Simple Sequence Repeats (SSR) PRINCIPLE
The Simple Sequence Repeats (SSRs) or microsatellites
occur ubiquitously in all the prokaryotic and eukaryotic genomes. Dinucleotide
repeats like (CA)n and (GA)n are the most abundant
repeats in most of the eukaryotes (for eg. in humans (CA)n repeat
occurs once in every 30 kb). Change in number of repeats gives the length
polymorphism which is revealed by designing primers for the sequences flanking
the microsatellite repeat motif followed by PCR amplification and visualisation
in agarose or denaturing polyacrylamide gel. This process of amplification
and visualisation can also be automated by labelling the primers with fluorescent
dye. ADVANTAGES This method requires very small quantity of DNA (~5 ng/reaction) and it is PCR based. This method provides co-dominant and highly reproducible markers that can be readily shared between different labs. The whole microsatellite analysis is automated including multiplexing (analysis of multiple loci in the same lane using different fluorescent labelled primer sets) to enable the researchers to carry out large scale genetic mapping and population studies. Besides being excellent molecular markers for genetic mapping, microsatellite markers are very useful for population genetics, variety identification and protection, monitoring of seed purity and hybrid quality, gene tagging, germplasm evaluation and phylogenetic studies, studies of kinship, conservation genetics and forensics. LIMITATION 1. Analysis of SSRs requires prior characterisation of sequences flanking the repeats to allow the primer design for PCR amplification, which is experimentally quite labour intensive. 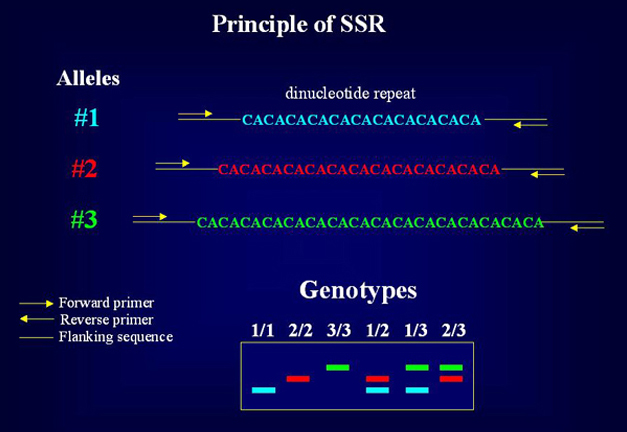
Click on the image to enlarge.
Protocols List:
 I.Protocol for analysing microsatellites using agarose gel electrophoresis 1. Equipment and reagents a) Thermal cycler b) Agarose gel electrophoresis system c) The reagents including Taq DNA polymerase, 10 x PCR buffer (500 mM KCl, 100 mM Tris-HCl, 0.01% gelatin and 1% Triton X-100), 10 x dNTPs stock (1 mM) and 10 ng DNA samples. d) 1 x TBE (90 mM Tris borate, pH 8.3, 2 mM EDTA) e) MetaPhor agarose (FMC) and normal agarose
2. SSR PCR reaction mix
3. Thermal Cycling Conditions
4. Agarose gel electrophoresis conditions Aliquots of amplified DNA from individual
PCR reactions should be loaded on a 2.5% (3 parts of Metaphor agarose:
1 part of agarose) gel in 1x TBE. Electrophoretic separations should
be performed in 1x TBE in a horizontal gel tank.  II. Protocol for analysing microsatellites using PAGE and silver staining 1. Equipment and reagents a) Thermal cycler b) Poly Acrylamide Gel Electrophoresis system c) The reagents including Taq DNA polymerase, 10 x PCR buffer (500 mM KCl, 100 mM Tris-HCl, 0.01% gelatin and 1% Triton X-100), 10 x dNTPs stock (1 mM) and 10 ng DNA samples. d) 1 x TBE (90 mM Tris borate, pH 8.3, 2 mM EDTA) e) Silver staining reagents like, fixative-stop solution (10% ethanol), silver nitrate solution (0.2% silver nitrate in water), developer solution (1.5% sodium hydroxide and 3ml/l formaldehyde).
2. SSR PCR reaction mix
3. Thermal Cycling Conditions
Aliquots of amplified DNA from individual
PCR reactions should be loaded on a denaturing 6% Poly Acrylamide Gel containing
7M urea in 1x TBE. After the bromophenol blue dye runs out, the gel must
be silver stained for band detection.
5. Silver staining After electrophoresis, gels should
be fixed in fixative-stop solution for 30 min. Fixed gel must be rinsed
3 times with water for 2 min. each. Later gel must be impregnated with silver
nitrate solution for 10 min. and rinsed with distilled water for 5-20 sec.
Gel can be developed with cold developer solution for 4 min. Developing
reaction should be stopped with fixative-stop solution for at least 1 min.
and washed extensively with water. The stained gels are dried at room temperature
and stored in photographic albums.  III. Protocol for analysing microsatellites using radiolabel 1. Equipment and reagents a) Thermal cycler b) Long gel Poly Acrylamide Gel Electrophoresis system c) Reagents including Taq DNA polymerase (preferably 'AmpliTaq Gold' to prevent stutter bands, see note below), 10 x PCR buffer (500 mM KCl, 100 mM Tris-HCl, and 1% Triton X-100), 10 x each of dGTP, dCTP dTTP and dATP stock (1 mM), a -32p dATP and 20 ng DNA samples. d) Stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue and 0.05% Xyline cyanol in the ratio of 3:2). e) 1 x TBE (90 mM Tris borate, pH 8.3, 2 mM EDTA)
2. SSR PCR reaction mix
3. Thermal Cycling Conditions
Aliquots of amplified DNA from individual
PCR reactions should be mixed with formamide stop solution. Four micro-litre
of the sample must be denatured at 95oC for 2 min., and immediately
chilled on ice. Electrophoretic separation must be done on 6% polyacrylamide
gel containing 8 M urea in 1 x TBE buffer. After electrophoresis, the gels
should be fixed for 2 x 20 min. with 10% glacial acetic acid. The fixed
gel must be air-dried and exposed for 4 -12 hrs.  IV. Protocol for analysing microsatellites using fluorescent dNTP** 1. Equipment and reagents a) Thermal cycler b) Automated sequencing System (e.g. ABI 377) c) The reagents including Taq DNA polymerase (preferably 'AampliTaq Gold' to prevent stutter bands, see Note below), 10 x PCR buffer (500 mM KCl, 100 mM Tris-HCl, and 1% Triton X-100), 10 x each of dGTP, dCTP dTTP and dATP stock (1 mM), 2m M stock of fluorescent dUTP (TAMARA, R110 or R6G, Perkin Elmer) and 5 ng DNA samples. d) 6 x loading buffer and GENESCAN-1000 ROX- labelled molecular weight standard e) 1 x TBE (90 mM Tris borate, pH 8.3, 2 mM EDTA)
3. Thermal Cycling Conditions
4. Sample preparation and electrophoresis conditions One micro-litre of PCR product should
be mixed with 1.5 m l of 6 x loading buffer (1:
4 mixture of loading buffer and formamide; Sigma). To this add 0.3 m l of GENESCAN-500 ROX- labelled molecular weight
standard (red fluorescence). Before loading onto an ABI 377 automated sequencer,
the samples should be denatured at 92° C for
1 min. For an optimum separation, denaturing Polyacrylamide gel of 5% containing
6M urea in 1 X TBE buffer must be used. *NOTE 1: This is a sample reaction condition. Annealing
temperature and MgCl2 concentration vary for each primer set. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Copyright © 2004 All Rights Reserved, CDFD, Hyderabad, India | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||