|
|
|
| Home » Transcription » Research |
| Transcription |
Current Research Interests
Laboratory of transcription is engaged in understanding the mechanism, physiology and inhibition of bacterial Rho-dependent termination. A wide range of techniques from biophysics (spectroscopy, thermodynamics, fast kinetics, etc.), biochemistry (protein purification, chemical and enzymatic foot-printing of protein and nucleic acids, cross-linking, etc.), molecular biology (recombinant DNA techniques, site-directed mutagenesis, super resolution microscopy), bacterial genetics and genomics are used in the laboratory to solve these intellectually challenging problems.
Projects:
- Mechanism of transcription termination by transcription termination factor Rho.
- Mechanism of Rho-NusG interaction in vivo and in vitro.
- Physiological roles of Rho-dependent terminations.
- Super-resolution microscopy of the transcription machinery.
- Fast-kinetics approach to study the transcription termination processes.
- Isolation of mycobacteriophage derived proteins with antimicrobial activies.
- Design of antimicrobial peptides from bacteriophage proteins.
Collaborators::
- Prof Markus Wahl, Freie Universität Berlin, Germany (Cryo-EM).
- Prof. Udaydittya Sen, SINP, Kolkata (Crystallography).
- Prof. Agnieszka Szalewska-Palas, Uniwersytet Gdanski, Poland.
Research Highlights |
|
In vivo regulation of bacterial Rho-dependent transcription termination by the nascent RNA (JBC 2022, in press).
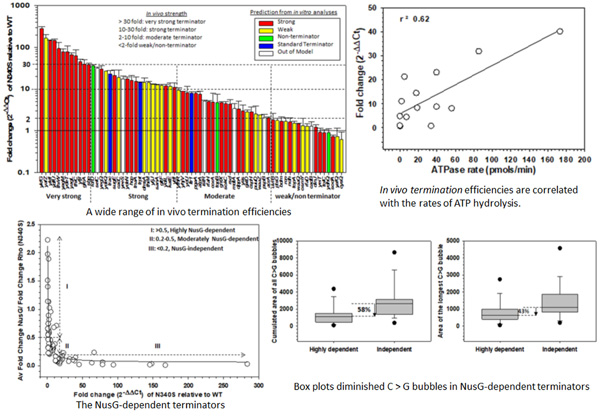
Bacterial Rho is a RNA-dependent ATPase that functions in the termination of DNA transcription. However, the in vivo nature of the bacterial Rho-dependent terminators, as well as the mechanism of the Rho-dependent termination process, are not fully understood. Here, we measured the in vivo termination efficiencies of 72 Rho-dependent terminators in E.coli by systematically performing qRT-PCR analyses of cDNA prepared from mid-log phase bacterial cultures. We found that these terminators exhibited a wide range of efficiencies, and many behaved differently in vivo compared to the predicted or experimentally determined efficiencies in vitro. Rho-utilization sites (rut sites) present in the RNA terminator sequences are characterized by the presence of C-rich/G-poor sequences, or C>G bubbles. We found that weaker terminators exhibited a robust correlation with the properties (size, length, density, etc.) of these C>G bubbles of their respective rut sites, while stronger terminators lack this correlation, suggesting a limited role of rut sequences in controlling in vivo termination efficiencies. We also found that in vivo termination efficiencies are dependent on the rates of ATP hydrolysis as well as Rho-translocation on the nascent RNA. We demonstrate that weaker terminators, in addition to having rut sites with diminished C>G bubble sizes, are dependent on the Rho-auxiliary factor, NusG, in vivo. From these results, we concluded that in vivo Rho-dependent termination follows a nascent RNA-dependent pathway, where Rho-translocation along the RNA is essential and rut sequences may recruit Rho in vivo, but Rho-rut binding strengths do not regulate termination efficiencies.
|
|
Rho-dependent transcription termination regulates the toxin–antitoxin modules of cryptic prophages to silence their expression in Escherichia coli (FEBS letters, 2021).
Bacterial Rho-dependent transcription termination regulates many physiological processes. Here, we report that it controls the expression of toxin–antitoxin (TA) modules of cryptic prophages in E. coli. Microarray profiles of Rho mutants showed upregulation of genes of the CP4-6 and CP4-44 prophages, including their TA modules, that were validated by RT-qPCR. Analysis of the in vivo termination efficiency and the mRNA sequences of these prophages revealed the presence of many Rho-dependent terminators. The prophage TA modules exhibited synthetic lethality with the Rho mutants, indicating functional involvement of Rho-dependent termination in controlling these modules. Rho-dependent termination does not regulate most of the chromosomal TA modules. We conclude that Rho-dependent termination specifically silences the TA modules of prophages, thereby augmenting bacterial innate immunity.
|
|
Design of novel peptide-inhibitors against the conserved bacterial transcription terminator, Rho (JBC, 2021).
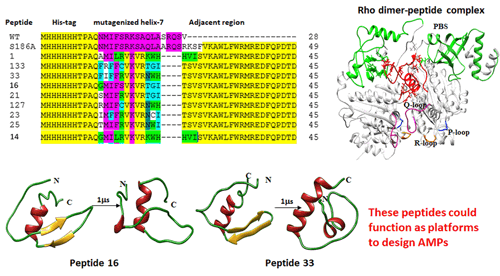
The transcription terminator Rho regulates many physiological processes in bacteria, such as antibiotic sensitivity, DNA repair, RNA-remodeling, etc, and hence, is a potential antimicrobial target, which is unexplored. The bacteriophage P4 capsid protein, Psu, moonlights as a natural Rho antagonist. Here, we report the design of novel peptides based on the C-terminal region of Psu using phenotypic screening methods. The resultant 38-mer peptides, in addition to containing mutagenized Psu sequences, also contained plasmid sequences, fused to their C-termini. Expression of these peptides inhibited the growth of E. coli, and specifically inhibited Rho-dependent termination in vivo. Peptides 16 and 33 exhibited the best Rho-inhibitory properties in vivo. Direct high-affinity binding of these two peptides to Rho also inhibited the latter's RNA-dependent ATPase and transcription termination functions in vitro. These two peptides remained functional even if 8-10 amino acids were deleted from their C-termini. In-silico modeling and genetic and biochemical evidence revealed that these two peptides bind to the primary RNA binding site of the Rho hexamer near its subunit interfaces. Additionally, the gene expression profiles of these peptides and Psu overlapped significantly. These peptides also inhibited the growth of Mycobacteria, and inhibited the activities of Rho proteins from M. tuberculosis, Xanthomonas, V. cholerae, and S. enterica. Our results showed that these novel anti-Rho peptides mimic the Rho-inhibition function of the ~42 kDa dimeric bacteriophage P4 capsid protein, Psu. We conclude that these peptides and their C-terminal deletion derivatives could provide a basis on which to design novel anti-microbial peptides (AMP).
|
|
Projects in progress:
- Understanding the physiological consequences of Rho-dependent termination.
- Understanding the role of the omega subunit of RNAP in Rho-dependent termination.
- In vivo localization of Rho and NusG by super-resolution microscopy.
- Identification Rho-RNAP functional interaction domains.
- Isolation and characterization of anti-mycobacterial proteins from mycobacteriophages.
- Design of antiterminator peptides from Psu protein.
Extramural Funding:
- SERB JC Fellowship (2022-2027)
- DST-Indo-Polish Grant (2021-2022)
- Indo-German ICMR grant (2020-2023).
- DST-SERB Grant (2020-2023).
Awards/Recognition:
- 2002-2007: GRIP research grant award from NIH, USA.
- 2003-2008: Wellcome Trust, UK, Senior Research Fellowship.
- 2007: DBT Bioscience carrier development award.
- 2007: Elected member of GRC.
- 2008: DST Swarnajayanti Research Fellowship.
- 2011: Elected fellow of NASI, Allahabad.
- 2015: Member DST- SERB, task force.
- 2018: Elected Fellow INSA, New Delhi.
- 2018: Elected fellow IASc, Bangalore.
- 2018: Fellow of Telangana Academy of Sciences.
- 2019: DBT TATA Innovation Fellowship.
- 2021: Elected Fellow of West Bengal Academy of Sciences.
- 2022: SERB JC Bose Fellowship.
Editorial Board Member:
- Journal of Applied Genetics (microbial Genetics section).
Reviewer of Journals/grants/Thesis:
- Nature Communications, MBio, TIBS, Journal of Bacteriology, Journal of Molecular Biology, Microbiology, PLOS one, Biochemical transactions, Communication Biology, msphere, MEEGID, Microbial Genomics, Scientific Reports, Indian Journal of Biophysics and Biochemistry, Journal of Bioscience, etc.
- The reviewer of grants for different granting agencies like DBT, DST, DRDO, Indo-French programs, Marsden Fund, New Zealand. etc.
- The reviewer of the thesis of the Ph.D. students from renowned institutions like SINP, Kolkata; Bose Institute, Kolkata; IISc., Bangalore; IMTech, Chandigarh, HCU, JNU, etc.
Patents:
- 'NOVEL SYNTHETIC PEPTIDES'; Indian Patent Application No. 201841048582 filed on December 20, 2019.
| |
|
|
|
| Last updated on : Friday, 29th April, 2022. |
|
|
|
|